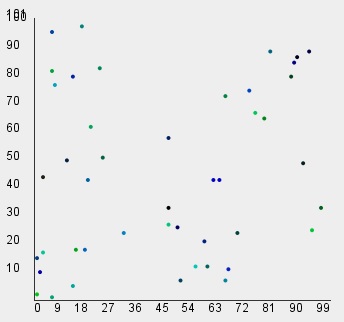
ClusteringLib clib=new ClusteringLib();
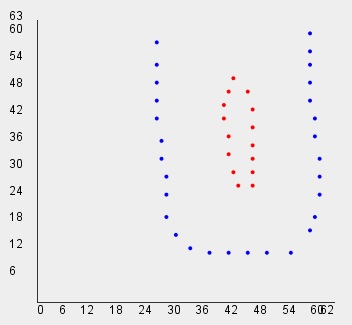
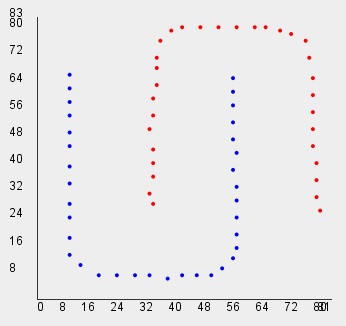
// Read U-shape dataset for Shape Independent Clustering
int[][] data=vlib.readDST_2D_int("u_shape");
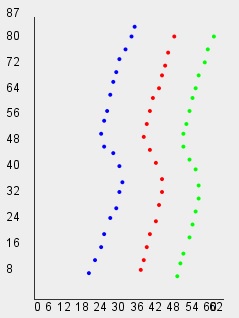
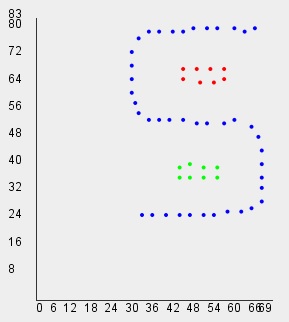
// Shape Independent Clustering with default threshold=1.5
int[] clusters=clib.ShapeIndependentClustering(data);
vlib.setDraw(data, clusters);